pcdna3 1 map
Related Articles: pcdna3 1 map
Introduction
In this auspicious occasion, we are delighted to delve into the intriguing topic related to pcdna3 1 map. Let’s weave interesting information and offer fresh perspectives to the readers.
Table of Content
- 1 Related Articles: pcdna3 1 map
- 2 Introduction
- 3 Unveiling the Power of pcDNA3.1: A Comprehensive Guide to the Vector
- 3.1 Understanding the Blueprint: A Detailed Look at pcDNA3.1
- 3.2 Beyond the Structure: The Functional Capabilities of pcDNA3.1
- 3.3 Navigating the Applications: Diverse Use Cases of pcDNA3.1
- 3.4 FAQs: Addressing Common Questions
- 3.5 Tips for Success: Maximizing the Potential of pcDNA3.1
- 3.6 Conclusion: A Powerful Tool for Modern Research
- 4 Closure
Unveiling the Power of pcDNA3.1: A Comprehensive Guide to the Vector

The realm of molecular biology is built upon the foundation of vectors, tools that serve as vehicles for carrying and delivering genetic material into cells. Among these, pcDNA3.1 stands out as a versatile and widely used expression vector, facilitating the efficient expression of genes of interest in various cell lines. This comprehensive guide delves into the intricacies of pcDNA3.1, exploring its structure, functionalities, and applications, ultimately highlighting its significance in modern research.
Understanding the Blueprint: A Detailed Look at pcDNA3.1
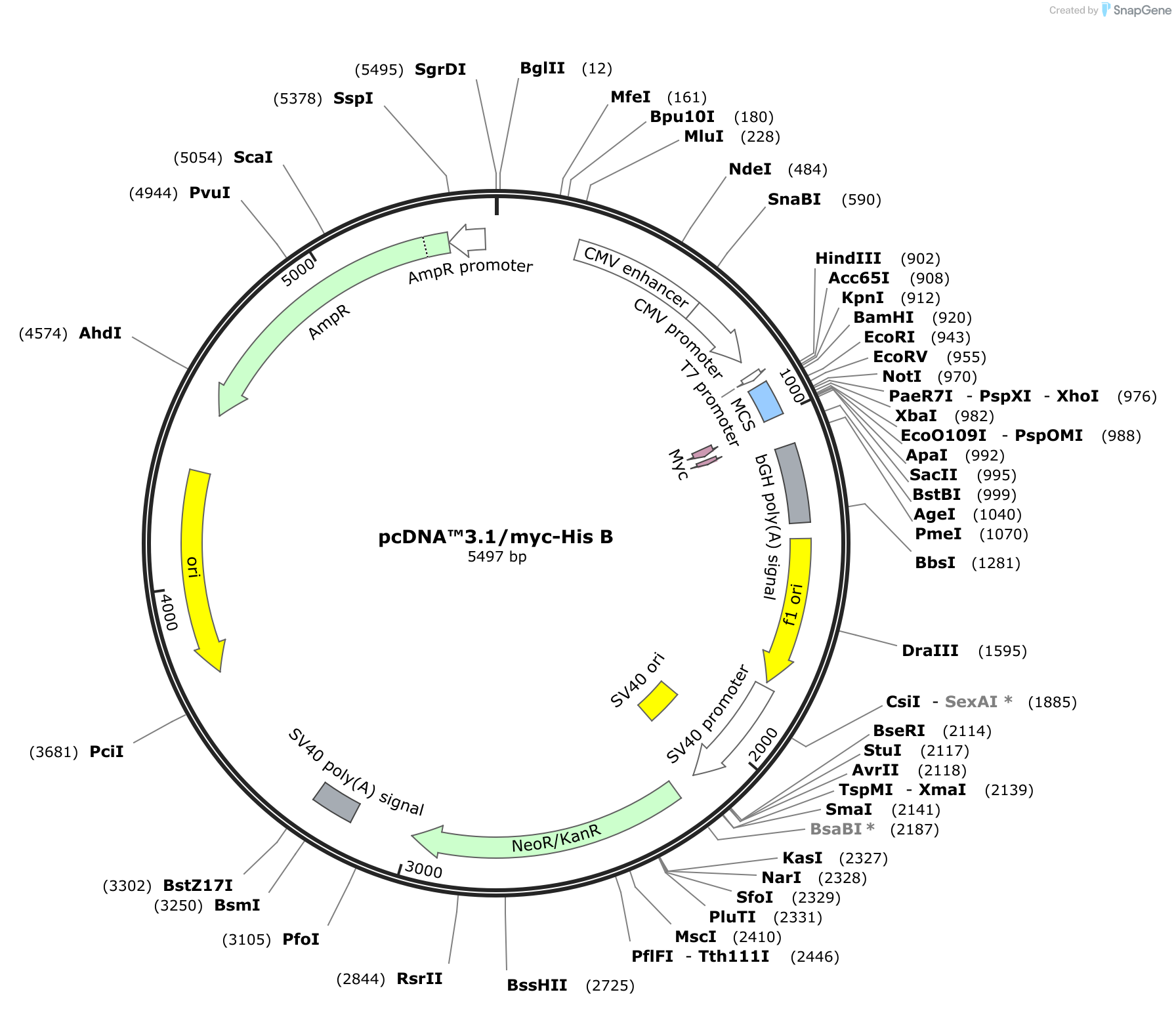
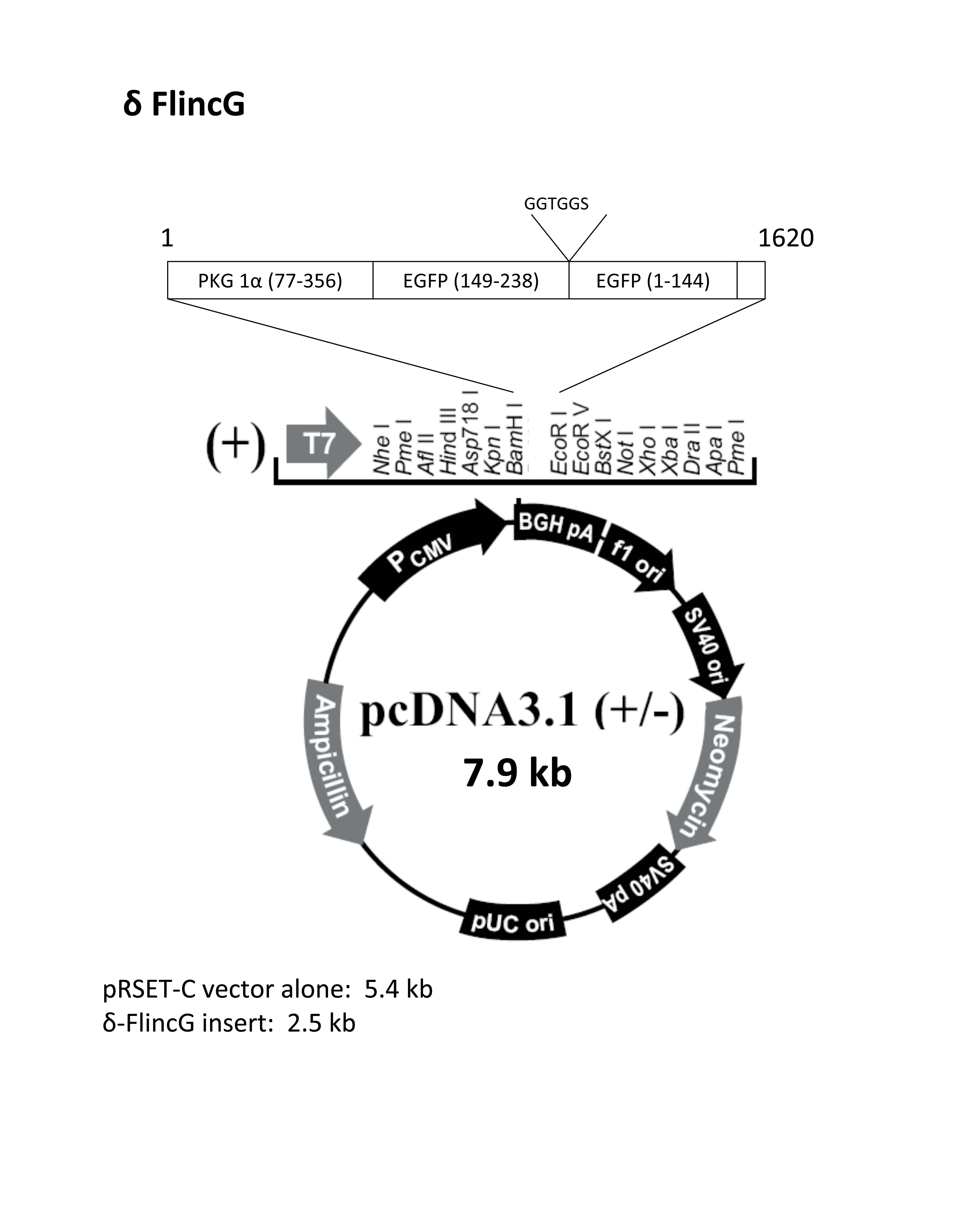
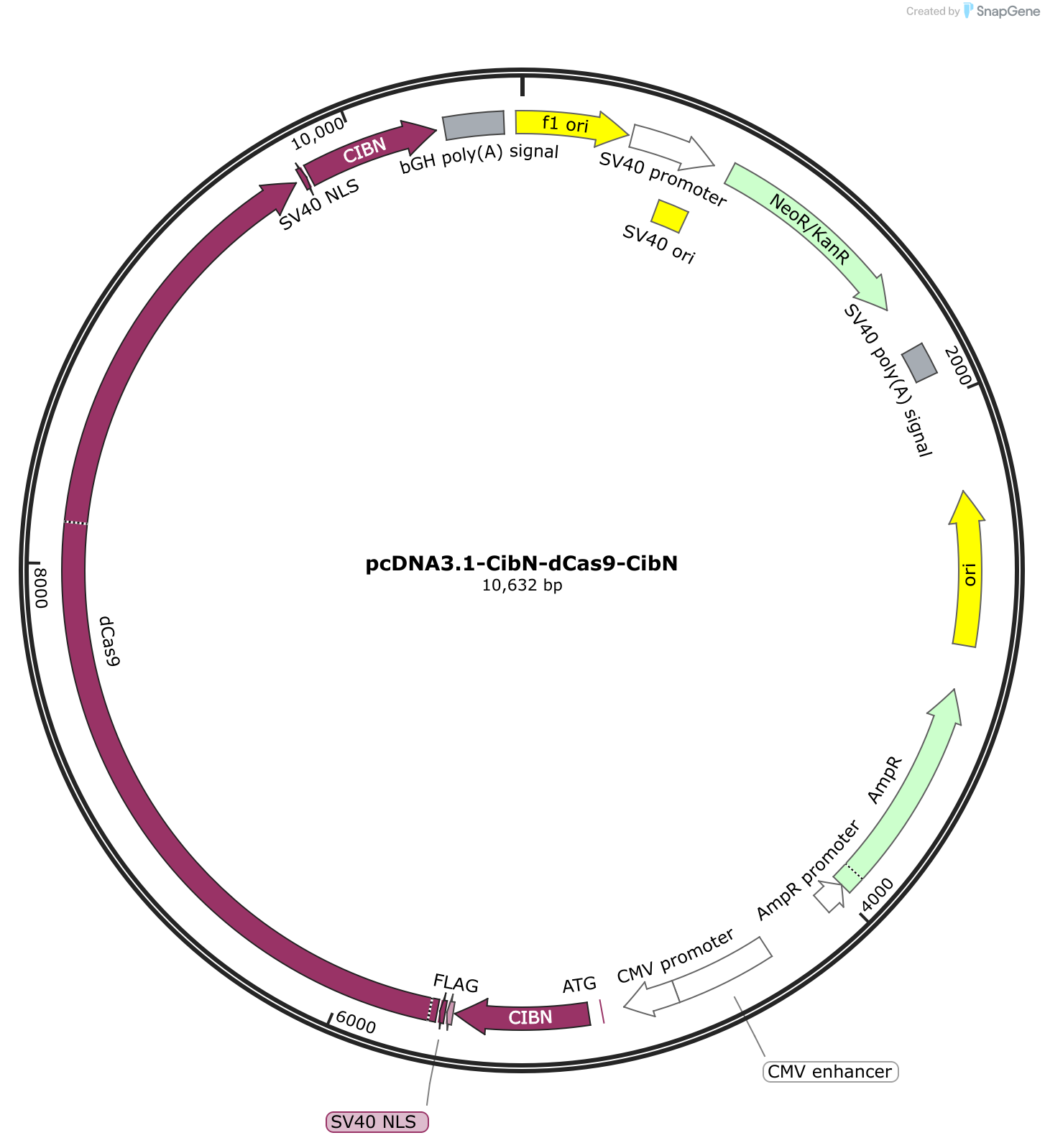
pcDNA3.1, a member of the pcDNA family, is a robust plasmid vector meticulously engineered for high-level gene expression in mammalian cells. It is comprised of several essential components, each playing a crucial role in its functionality:
1. Origin of Replication (ori): This sequence serves as the starting point for DNA replication within the host cell. pcDNA3.1 employs the SV40 origin of replication, ensuring efficient replication within mammalian cells.
2. Promoter Region: The promoter sequence is responsible for initiating gene transcription. pcDNA3.1 utilizes the powerful CMV (cytomegalovirus) promoter, known for its high level of expression in a wide range of mammalian cells.
3. Multiple Cloning Site (MCS): The MCS is a strategically designed region containing a series of unique restriction enzyme recognition sites. This allows researchers to easily insert their gene of interest into the vector, placing it under the control of the CMV promoter.
4. Selection Markers: These genetic elements are essential for identifying cells that have successfully incorporated the pcDNA3.1 vector. pcDNA3.1 features two selection markers:
* **Neomycin Resistance Gene (neo):** This gene confers resistance to the antibiotic G418, allowing for the selection of cells that have integrated the vector into their genome.
* **Ampicillin Resistance Gene (amp):** This gene provides resistance to the antibiotic ampicillin, enabling the selection of bacterial cells containing the plasmid during propagation.5. Polyadenylation Signal (pA): This sequence is crucial for proper mRNA processing and stability. It signals the addition of a poly-A tail to the transcribed mRNA, enhancing its stability and promoting efficient translation.
6. SV40 Enhancer: This element further enhances gene expression by increasing the rate of transcription initiation.
Beyond the Structure: The Functional Capabilities of pcDNA3.1
The intricate design of pcDNA3.1 translates into a range of capabilities, making it a valuable tool for diverse research applications:
1. Gene Expression and Analysis: pcDNA3.1 serves as a robust platform for expressing genes of interest in mammalian cells. Its strong CMV promoter ensures high levels of protein production, facilitating downstream analyses such as protein function studies, antibody production, and drug screening.
2. Functional Studies: By introducing modified genes or specific mutations into cells, researchers can investigate the impact of these alterations on cellular processes. pcDNA3.1 provides a reliable tool for studying gene function, unraveling the intricacies of cellular pathways, and understanding disease mechanisms.
3. Protein Production: pcDNA3.1 is widely used for producing recombinant proteins. Its high-level expression capabilities allow for the production of large quantities of proteins for various applications, including therapeutic development, antibody generation, and structural studies.
4. Cell Line Engineering: pcDNA3.1 can be used to generate stable cell lines expressing specific genes. This enables researchers to create cell lines with desired phenotypes, providing valuable models for studying cellular processes, drug responses, and disease progression.
5. Therapeutic Development: pcDNA3.1 plays a crucial role in gene therapy research, serving as a vector for delivering therapeutic genes to target cells. This approach holds promise for treating genetic disorders and developing novel therapeutic strategies.
Navigating the Applications: Diverse Use Cases of pcDNA3.1
The versatility of pcDNA3.1 has led to its widespread adoption across various research fields:
1. Biomedical Research: pcDNA3.1 is extensively used for studying gene function, disease mechanisms, and drug discovery. Researchers utilize it to express specific genes, investigate protein interactions, and screen for potential therapeutic compounds.
2. Biotechnology and Pharmaceutical Industry: pcDNA3.1 is a key tool for protein production, enabling the development of therapeutic proteins, antibodies, and other biopharmaceuticals. Its high-level expression capabilities make it ideal for large-scale production.
3. Genetic Engineering and Synthetic Biology: pcDNA3.1 facilitates the creation of genetically modified organisms, enabling the development of novel crops, biofuels, and other biotechnological applications.
4. Fundamental Biological Research: pcDNA3.1 is a valuable tool for studying cellular processes, gene regulation, and protein function. Its high expression levels and ease of use make it a versatile instrument for fundamental research.
FAQs: Addressing Common Questions
1. What is the difference between pcDNA3.1 and pcDNA3.1(+) ?
pcDNA3.1(+) is a modified version of pcDNA3.1 containing a multiple cloning site (MCS) downstream of the CMV promoter, while pcDNA3.1 has the MCS upstream of the CMV promoter. This difference in MCS placement can affect the efficiency of gene expression and protein production.
2. How can I choose the appropriate pcDNA3.1 vector for my research?
The choice of pcDNA3.1 vector depends on the specific requirements of your research. Consider factors such as the desired expression level, the target cell line, and the presence of additional features like fluorescent tags or inducible promoters.
3. How can I insert my gene of interest into pcDNA3.1?
Gene insertion into pcDNA3.1 is typically achieved through restriction enzyme digestion and ligation. The gene of interest is first amplified using PCR, incorporating restriction sites that match those in the pcDNA3.1 MCS. Both the vector and the gene fragment are then digested with the appropriate restriction enzymes, followed by ligation to generate the recombinant vector.
4. How can I select for cells that have incorporated pcDNA3.1?
pcDNA3.1 contains the neomycin resistance gene (neo), which confers resistance to the antibiotic G418. Cells that have successfully integrated the vector into their genome will be resistant to G418, allowing for their selection.
5. What are the limitations of pcDNA3.1?
While pcDNA3.1 is a versatile vector, it has some limitations. It is primarily designed for mammalian cells, and its expression levels can vary depending on the target cell line. Additionally, its size can pose challenges for efficient delivery into certain cell types.
Tips for Success: Maximizing the Potential of pcDNA3.1
1. Optimize Gene Expression: Experiment with different promoter strengths, expression cassettes, and cell lines to achieve optimal gene expression levels.
2. Validate Gene Insertion: Confirm the successful insertion of your gene of interest into pcDNA3.1 by sequencing or restriction enzyme digestion.
3. Verify Protein Expression: Use Western blotting or other techniques to confirm the expression of your protein of interest.
4. Control for Background Expression: Include a control group with an empty vector to assess background expression levels and ensure that observed effects are due to the expression of your gene of interest.
5. Use Appropriate Selection Methods: Choose the appropriate selection marker and antibiotic for your experiment to ensure the isolation of cells that have integrated the pcDNA3.1 vector.
Conclusion: A Powerful Tool for Modern Research
pcDNA3.1 has emerged as a cornerstone in molecular biology research, providing a robust platform for gene expression, functional studies, and protein production. Its versatile design, combined with its high-level expression capabilities, has made it an indispensable tool across diverse fields, from biomedical research to biotechnology and beyond. By understanding the intricacies of pcDNA3.1 and following best practices, researchers can harness its power to advance scientific knowledge and develop innovative solutions for human health and well-being.
/pcDNA3.1(-).png)




Closure
Thus, we hope this article has provided valuable insights into pcdna3 1 map. We appreciate your attention to our article. See you in our next article!